Has AlphaFold solved the protein folding problem of biology?
As people around the world marveled at the most detailed images of the cosmos taken by the James Webb Space Telescope, biologists saw for the first time a different set of images, one that could help revolutionize life science research.
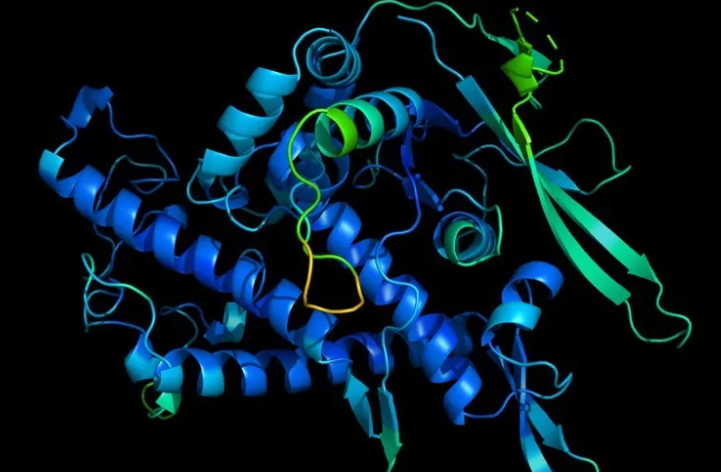
The images are the predicted three-dimensional shapes of more than 200 million proteins, rendered by an artificial intelligence system called AlphaFold, arguably covering the entire protein universe. Combining various deep learning techniques, the computer program is trained to predict protein shapes by recognizing patterns in structures that have already been resolved through decades of experimental work using electron microscopes and other methods.

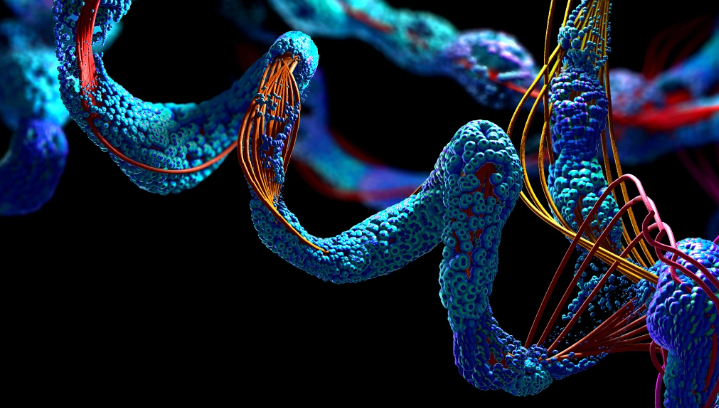
The first AI breakthrough came in 2021, with predictions for 350,000 protein structures, including nearly all known human proteins, you can look up a three-dimensional structure of a protein almost as easily as doing a Google keyword search. Researchers have used some of the predictions to develop potential new malaria vaccines, improve understanding of Parkinson’s disease, figure out how to protect bee health, gain insight into human evolution, and more.
The architecture of a protein is more than just aesthetics; can determine how that protein works, for example, proteins called enzymes need a pocket where they can capture small molecules and carry out chemical reactions and proteins that work in a protein complex (two or more proteins that interact like parts of a machine ) need the correct shapes to fit into the formation with their partners. Knowing the folds, coils, and loops in a protein’s shape can help scientists figure out how, for example, a mutation alters that shape to cause disease. That knowledge could also help researchers make better vaccines and drugs.
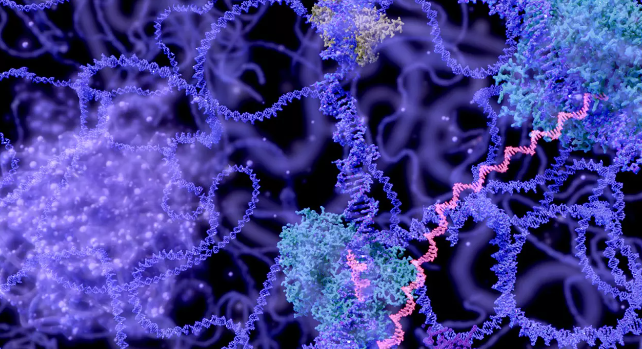
Some researchers, for example, have used AlphaFold’s predictions to help bring them closer to completing a huge biological puzzle: the nuclear pore complex. Nuclear pores are key portals that allow molecules to enter and exit cell nuclei, without the pores cells do not function properly. Each pore is huge, relatively speaking, made up of about 1,000 pieces of about 30 different proteins. Computer representations give biologists an advantage in solving problems such as how a drug might interact with a protein.


Responses